crAssphage
Overview
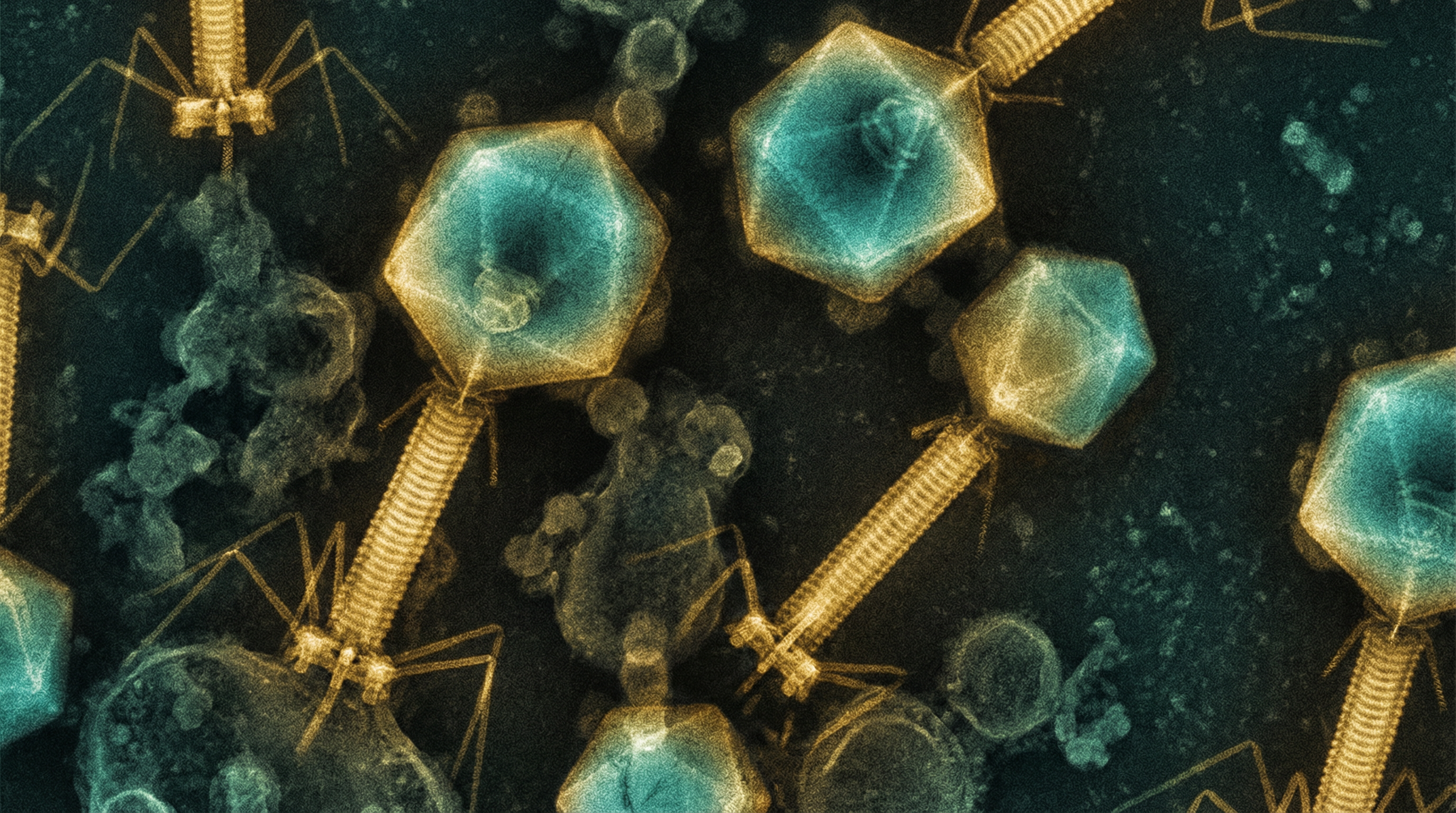
crAssphage (cross-assembly phage) represents the most abundant bacteriophage family in the human gut microbiome. It was discovered in 2014 through computational analysis of human fecal metagenomes, where researchers found highly abundant sequences that, when cross-assembled from multiple sources, indicated the presence of a unique bacteriophage. Despite being found in approximately 50% of individuals worldwide and representing up to 90% of human gut viromes in some subjects, crAssphage remained uncultured and understudied until recently. In 2018, researchers successfully isolated a crAssphage (ΦCrAss001) from human fecal material, confirming that it infects the human gut symbiont Bacteroides intestinalis.
The name "crAssphage" derives from the cross-assembly (crAss) technique used in its discovery. Since then, an entire family of related viruses, termed crAss-like bacteriophages, has been identified in various environments, including the human gut, termite gut, terrestrial/groundwater, and oceans.
Characteristics
crAssphage belongs to a diverse family of crAss-like bacteriophages with the following key characteristics:
- Genome: Double-stranded DNA (dsDNA) with a circular or circularly permuted genome of approximately 97-102 kb
- Morphology: Podovirus-like structure with an icosahedral capsid and short tail
- Taxonomy: Recently elevated to its own viral order, Crassvirales, belonging to the viral class Caudoviricetes
- Host Range: Primarily infects bacteria of the order Bacteroidales, particularly Bacteroides intestinalis and other Bacteroides species
- Replication Strategy: Unusual ability to replicate without disrupting host bacterial proliferation, maintaining stable co-existence
- Genomic Features: Contains approximately 105 protein-coding genes and multiple tRNA genes
The genome of crAssphage has interesting structural features, including its apparent division into two parts of roughly equal size with strictly opposite gene orientation and inverted GC skew, possibly reflecting the direction of transcription and/or replication. Despite the absence of obvious lysogeny genes, crAssphage replicates in a way that does not disrupt proliferation of the host bacterium and can maintain itself in continuous host culture for several weeks.
Role in Human Microbiome
crAssphage is a dominant component of the human gut virome, representing a significant portion of the viral community in the gastrointestinal tract. Its prevalence and abundance suggest it plays an important role in shaping the bacterial community structure and function in the human gut.
As a bacteriophage that infects Bacteroides species, which are among the most abundant bacteria in the human gut, crAssphage likely contributes to the regulation of bacterial population dynamics. This regulation may have downstream effects on gut homeostasis, nutrient cycling, and potentially host health.
The ability of crAssphage to maintain stable colonization of the mammalian gut without causing rapid lysis of its bacterial host suggests a complex relationship that may have evolved to promote coexistence rather than elimination of the host bacteria. This type of relationship could contribute to the overall stability of the gut microbiome.
Health Implications
The direct health implications of crAssphage are still being investigated, but several potential roles have been proposed:
Microbiome Modulation
As a regulator of Bacteroides populations, crAssphage may indirectly influence gut health by maintaining the balance of this important bacterial group. Bacteroides species are involved in various beneficial functions, including the breakdown of complex carbohydrates and the production of short-chain fatty acids.
Disease Associations
Changes in crAssphage abundance have been observed in various disease states, including inflammatory bowel disease (IBD). However, it remains unclear whether these changes are a cause or consequence of the disease. The relationship between crAssphage dynamics and human health is an active area of research.
Biomarker Potential
Due to its high specificity for human fecal contamination, crAssphage has been proposed as a valuable biomarker for water quality monitoring and environmental health assessment. Its presence in water sources can indicate human fecal pollution more specifically than traditional indicators.
Metabolic Activities
As a virus, crAssphage does not have its own metabolism but relies on hijacking the metabolic machinery of its bacterial host. However, the genome of crAssphage encodes several proteins that may influence the metabolism of its host:
Auxiliary Metabolic Genes: Some crAssphage genomes contain genes like β-fructosidase and ferredoxin-thioredoxin reductase, which may be involved in auxiliary metabolic processes unrelated to bacteriophage replication and virion assembly.
Nutrient Cycling: By lysing Bacteroides cells (even if at a slow rate), crAssphage may contribute to nutrient cycling in the gut ecosystem, releasing cellular contents that can be utilized by other microorganisms.
Horizontal Gene Transfer: Like other bacteriophages, crAssphage may facilitate horizontal gene transfer between bacterial hosts, potentially transferring metabolic genes or other functional elements.
Clinical Relevance
While crAssphage itself is not directly associated with any specific disease, its potential as a biomarker and its role in modulating the gut microbiome have clinical implications:
Microbiome-Based Diagnostics: Changes in crAssphage abundance or diversity could potentially serve as markers for gut dysbiosis or specific disease states.
Phage Therapy: Understanding the biology of crAssphage and its interaction with Bacteroides hosts could inform the development of phage-based therapies targeting pathogenic bacteria.
Environmental Monitoring: The high specificity of crAssphage for human fecal contamination makes it a valuable tool for monitoring water quality and environmental health.
Interactions with Other Microorganisms
crAssphage primarily interacts with its bacterial hosts, particularly Bacteroides intestinalis and other Bacteroides species. These interactions include:
Host Specificity: crAssphage shows specificity for bacteria of the order Bacteroidales, with Bacteroides intestinalis confirmed as a host through laboratory isolation.
Coevolution: The widespread distribution and abundance of crAssphage suggest a long history of coevolution with its bacterial hosts in the human gut.
Stable Coexistence: Unlike many virulent bacteriophages that rapidly lyse their hosts, crAssphage can maintain a stable relationship with its host bacteria, suggesting a more complex interaction that may benefit both parties.
Ecosystem Impact: By modulating Bacteroides populations, crAssphage may indirectly influence the broader microbial community structure and function in the gut.
Research Significance
crAssphage has significant research importance for several reasons:
Abundance and Prevalence: As the most abundant virus in the human gut, understanding crAssphage biology is crucial for comprehending the overall gut virome.
Model System: The recent isolation of crAssphage provides a valuable model system for studying phage-bacteria interactions in the gut.
Taxonomic Expansion: The discovery of crAssphage has led to the identification of an entire family of related viruses (crAss-like bacteriophages) and the establishment of a new viral order (Crassvirales).
Methodological Advances: The computational discovery of crAssphage highlighted the power of metagenomic approaches for identifying novel viruses, even when they cannot be cultured using traditional methods.
Biomarker Development: The specificity of crAssphage for human fecal contamination has led to its development as a biomarker for environmental monitoring.
Ongoing research aims to further elucidate the biology of crAssphage, its interaction with bacterial hosts, and its potential impact on human health and disease.