Escherichia coli
Escherichia coli is a gram-negative, facultatively anaerobic, rod-shaped bacterium that is one of the most well-studied microorganisms on Earth. First isolated and described by Theodor Escherich in 1885, E. coli has served as a model organism for countless scientific discoveries and is a ubiquitous member of the human gut microbiome. While primarily known as a commensal organism, certain pathogenic strains can cause a variety of intestinal and extraintestinal diseases.
Key Characteristics
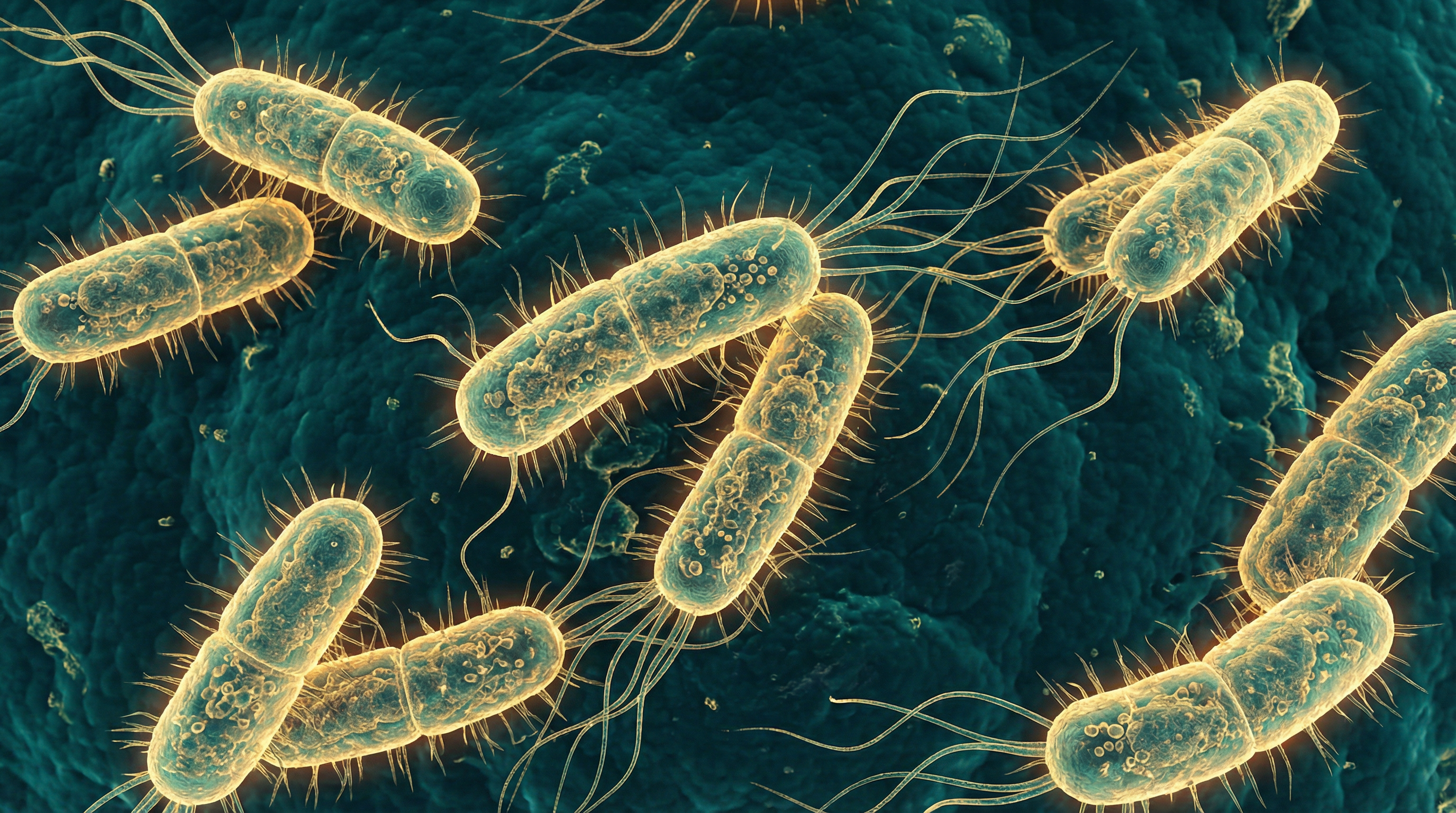
E. coli belongs to the family Enterobacteriaceae and the phylum Proteobacteria. It is a versatile bacterium with remarkable genetic diversity, which contributes to its ability to thrive in various environments and its diverse roles in human health and disease.
Morphologically, E. coli appears as a straight rod, approximately 2.0 μm long and 0.25–1.0 μm in diameter. It typically does not form spores and may be motile with peritrichous flagella or non-motile, depending on the strain. When grown on standard laboratory media, E. coli forms smooth, circular colonies that are often slightly convex with entire margins.
E. coli is a facultative anaerobe, meaning it can grow in both aerobic and anaerobic conditions. This metabolic versatility allows it to thrive in the oxygen-depleted environment of the gut as well as in more oxygenated regions near the mucosal surface. It can utilize a wide range of carbon sources and can grow on simple defined media with glucose as the sole carbon and energy source.
The species is genetically diverse, with a core genome of approximately 3,000 genes and a pan-genome (the total gene repertoire of the species) estimated to contain more than 20,000 genes. This genetic diversity underlies the remarkable adaptability of E. coli and its ability to occupy various ecological niches.
E. coli strains are often classified into phylogenetic groups (A, B1, B2, C, D, E, F, and G), with groups B2 and D containing most of the extraintestinal pathogenic strains, while commensal strains are more commonly found in groups A and B1.
Role in Human Microbiome
E. coli is one of the first colonizers of the human gut, typically establishing itself within days after birth. In the adult human gut, E. coli is present in over 90% of individuals, although it constitutes only about 0.1% of the total gut microbiota by abundance. Despite its relatively low abundance, E. coli plays several important roles in the gut ecosystem.
As a facultative anaerobe, E. coli is thought to help create and maintain the anaerobic environment necessary for the growth of obligate anaerobes by consuming oxygen at the mucosal surface. This "oxygen-scavenging" function may be particularly important during the early colonization of the infant gut, helping to establish the conditions needed for the development of a diverse microbiome.
Studies of E. coli population dynamics in the human gut have revealed both long-lived resident strains and short-lived transient strains. Resident strains can persist in the gut for months or even years, while transient strains may only be detected for days or weeks. Interestingly, these resident and transient strains often belong to different phylogenetic groups, suggesting that certain genetic characteristics may predispose some strains to long-term colonization.
The distribution of E. coli in the gut is not uniform. It is more abundant in the cecum and proximal colon and decreases in abundance toward the distal colon. It is also more abundant in the mucus layer adjacent to the epithelium than in the lumen, reflecting its adaptation to the mucosal environment.
Health Implications
Commensal Functions
As a commensal organism, E. coli contributes to several beneficial functions in the human gut:
Vitamin synthesis: E. coli can produce vitamin K2 (menaquinone), which is important for blood clotting and bone health. It may also contribute to the synthesis of certain B vitamins.
Colonization resistance: Commensal E. coli strains can help protect against enteric pathogens through various mechanisms, including competition for nutrients and attachment sites, production of inhibitory compounds, and stimulation of host immune responses.
Immune system development: Interaction with commensal E. coli helps train and regulate the host immune system, particularly in early life.
Metabolic functions: E. coli participates in various metabolic processes in the gut, including fermentation of carbohydrates and production of short-chain fatty acids, which serve as energy sources for colonocytes.
Recent research has demonstrated that certain commensal E. coli strains can be considered "health-promoting" due to their ability to protect against intestinal infections. For example, some strains have been shown to protect against Salmonella infection in mouse models through the production of specific metabolites.
Pathogenic Strains
While most E. coli strains are harmless or beneficial, certain pathogenic strains can cause a variety of diseases. These pathogenic strains are typically classified into several pathotypes based on their virulence mechanisms and the diseases they cause:
Enterohemorrhagic E. coli (EHEC): Also known as Shiga toxin-producing E. coli (STEC), these strains produce Shiga toxins that can cause bloody diarrhea and, in severe cases, hemolytic uremic syndrome (HUS), a potentially life-threatening condition characterized by kidney failure, thrombocytopenia, and hemolytic anemia. The most well-known EHEC serotype is O157:H7.
Enteropathogenic E. coli (EPEC): These strains cause watery diarrhea, particularly in infants and young children in developing countries. EPEC attaches to intestinal epithelial cells and causes characteristic "attaching and effacing" lesions.
Enterotoxigenic E. coli (ETEC): A major cause of traveler's diarrhea and a significant cause of diarrheal disease in children in developing countries. ETEC produces heat-labile and/or heat-stable enterotoxins that induce watery diarrhea.
Enteroaggregative E. coli (EAEC): Associated with persistent diarrhea in children and adults. EAEC adheres to intestinal epithelial cells in a characteristic "stacked-brick" pattern and produces various enterotoxins and cytotoxins.
Enteroinvasive E. coli (EIEC): Causes a dysentery-like illness similar to that caused by Shigella. EIEC invades and replicates within intestinal epithelial cells, causing inflammation and cell death.
Diffusely adherent E. coli (DAEC): Associated with diarrhea in children and urinary tract infections. DAEC adheres to epithelial cells in a diffuse pattern.
In addition to these intestinal pathotypes, certain E. coli strains can cause extraintestinal infections, including urinary tract infections (UTIs), meningitis, and sepsis. These extraintestinal pathogenic E. coli (ExPEC) strains typically belong to different phylogenetic groups than the intestinal pathotypes and possess specific virulence factors that enable them to colonize and cause disease outside the intestinal tract.
Associations with Other Conditions
Beyond its direct roles as a commensal or pathogen, E. coli has been associated with various other health conditions:
Inflammatory bowel disease (IBD): Some studies have found increased adherent-invasive E. coli (AIEC) in the intestinal mucosa of patients with Crohn's disease, suggesting a potential role in the pathogenesis of this form of IBD.
Colorectal cancer: Certain E. coli strains that produce colibactin, a genotoxin that can damage DNA, have been associated with colorectal cancer.
Metabolic disorders: Alterations in E. coli populations have been observed in conditions such as obesity and type 2 diabetes, although the causal relationships remain unclear.
Antibiotic resistance: E. coli can serve as a reservoir for antibiotic resistance genes in the gut microbiome, potentially contributing to the spread of resistance.
Metabolic Activities
E. coli exhibits diverse metabolic capabilities that enable it to thrive in the competitive environment of the human gut:
Carbohydrate metabolism: E. coli can utilize a wide range of carbohydrates, including glucose, lactose, and various other sugars. It preferentially uses glucose when available, but can switch to alternative carbon sources when necessary.
Mixed acid fermentation: Under anaerobic conditions, E. coli performs mixed acid fermentation, producing a mixture of products including acetate, formate, lactate, succinate, ethanol, CO2, and H2.
Respiration: In the presence of oxygen or alternative electron acceptors (such as nitrate), E. coli can perform respiration, which yields more energy than fermentation.
Amino acid metabolism: E. coli can synthesize all 20 common amino acids when necessary, but can also take up and metabolize amino acids from the environment.
Adaptation to nutrient limitation: E. coli has evolved sophisticated mechanisms to sense and respond to nutrient availability, allowing it to adapt to the changing conditions in the gut.
Biofilm formation: Many E. coli strains can form biofilms, which are structured communities of bacteria embedded in a self-produced matrix. Biofilm formation can enhance survival under adverse conditions and may contribute to colonization persistence.
These metabolic activities not only support the growth and survival of E. coli in the gut but also influence the broader gut ecosystem through the production of various metabolites and the consumption of resources.
Clinical Relevance
The clinical relevance of E. coli spans a wide spectrum, from its use as a probiotic to its role as a major human pathogen:
Probiotic applications: Certain non-pathogenic E. coli strains, such as E. coli Nissle 1917 (EcN), have been used as probiotics for the treatment of various gastrointestinal disorders, including ulcerative colitis, Crohn's disease, and irritable bowel syndrome. EcN has been shown to have anti-inflammatory effects, enhance barrier function, and provide colonization resistance against pathogens.
Diagnostic indicator: The presence of E. coli in water or food is used as an indicator of fecal contamination, which may signal the potential presence of enteric pathogens.
Pathogen detection and identification: Various methods have been developed to detect and identify pathogenic E. coli strains in clinical samples, food, and water. These include culture-based methods, immunological assays, and molecular techniques such as PCR and whole-genome sequencing.
Antimicrobial resistance: E. coli is a major reservoir of antimicrobial resistance genes, and the prevalence of resistant strains has been increasing globally. Of particular concern are extended-spectrum beta-lactamase (ESBL)-producing and carbapenem-resistant E. coli, which limit treatment options for serious infections.
Vaccine development: Efforts are ongoing to develop vaccines against various pathogenic E. coli strains, particularly ETEC (a major cause of traveler's diarrhea) and ExPEC (which causes urinary tract infections and other extraintestinal infections).
Model organism: E. coli continues to serve as an important model organism for understanding basic biological processes, developing new biotechnologies, and testing novel antimicrobial strategies.
The clinical management of E. coli infections varies depending on the pathotype and the site of infection. While many uncomplicated infections resolve without specific treatment, severe or invasive infections may require antimicrobial therapy, supportive care, and in some cases, surgical intervention.
Interaction with Other Microorganisms
E. coli engages in complex interactions with other members of the gut microbiome:
Competition with other Enterobacteriaceae: E. coli competes with other members of the Enterobacteriaceae family, such as Klebsiella, Citrobacter, and Enterobacter, for similar niches and resources in the gut.
Relationship with anaerobes: E. coli has a complex relationship with obligate anaerobes in the gut. By consuming oxygen, E. coli helps create the anaerobic environment needed by these bacteria. In turn, some anaerobes produce metabolites that can be utilized by E. coli.
Bacteriocin production and sensitivity: Many E. coli strains produce bacteriocins, which are narrow-spectrum antimicrobial peptides that can inhibit the growth of closely related bacteria. This may contribute to competition among different E. coli strains and other Enterobacteriaceae.
Horizontal gene transfer: E. coli can exchange genetic material with other bacteria through various mechanisms of horizontal gene transfer, including conjugation, transformation, and transduction. This contributes to the genetic diversity of E. coli and the spread of traits such as antibiotic resistance and virulence factors.
Phage interactions: E. coli is susceptible to infection by numerous bacteriophages, which can influence its population dynamics and evolution. Some phages can transfer virulence genes between bacteria through the process of transduction.
Quorum sensing: E. coli can communicate with other bacteria through quorum sensing, a process by which bacteria detect and respond to population density through the production and detection of signaling molecules.
These interactions contribute to the complex ecology of the gut microbiome and may influence the colonization success and persistence of different E. coli strains.
Research Significance
E. coli has been and continues to be one of the most important model organisms in biological research:
Molecular biology: Many fundamental discoveries in molecular biology were made using E. coli, including the genetic code, DNA replication, transcription, translation, and gene regulation.
Genetics and genomics: E. coli has been instrumental in the development of genetic and genomic techniques, from early gene mapping to modern genome editing methods.
Synthetic biology: E. coli is a workhorse of synthetic biology, used for the design and construction of novel biological systems with practical applications.
Biotechnology: E. coli is widely used for the production of recombinant proteins, biofuels, and other valuable compounds.
Evolutionary biology: Studies of E. coli have provided insights into evolutionary processes, including adaptation, speciation, and the evolution of cooperation and conflict.
Microbiome research: As a common member of the human gut microbiom (Content truncated due to size limit. Use line ranges to read in chunks)