Candidatus Saccharibacteria
Key Characteristics
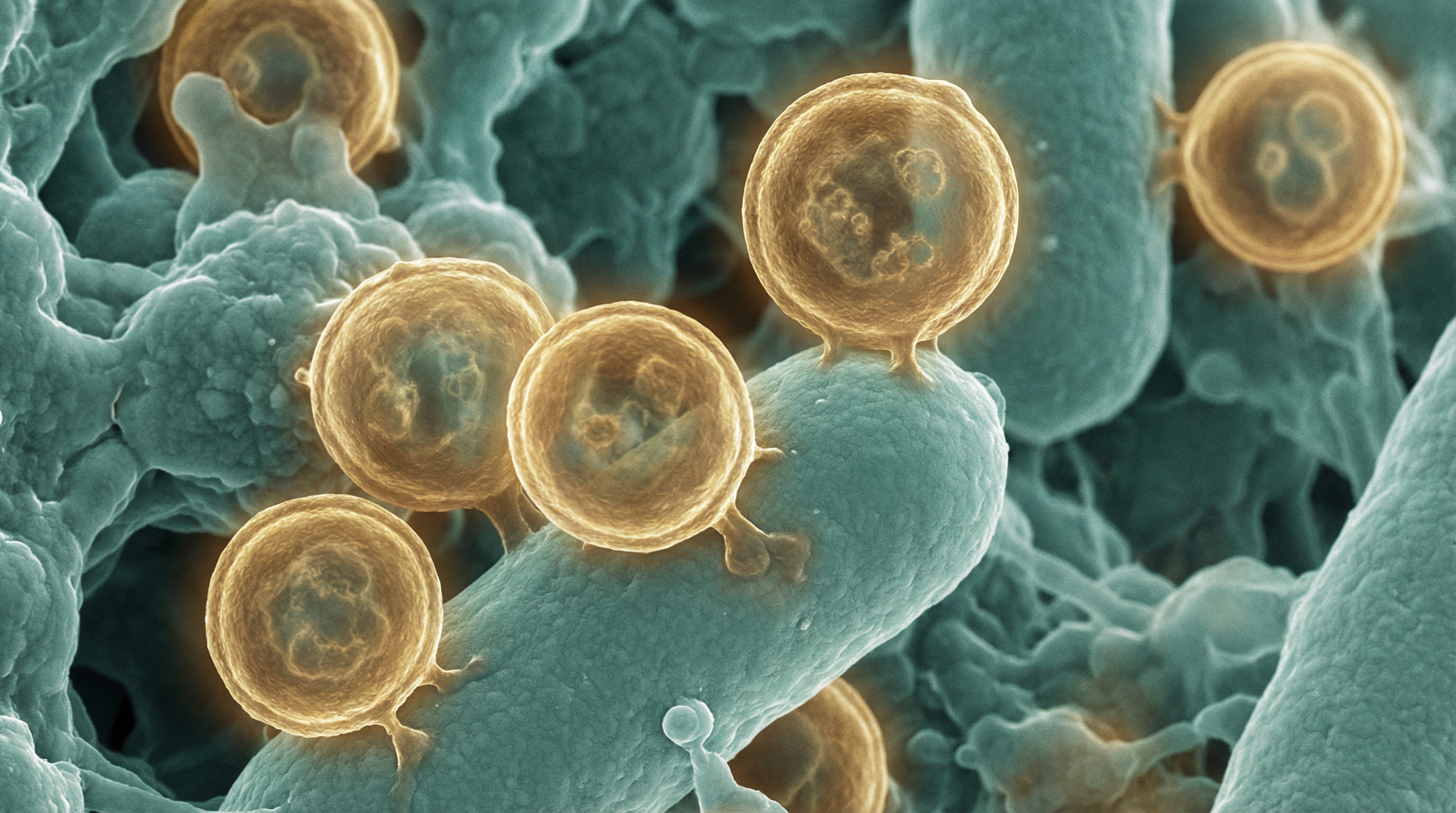
Candidatus Saccharibacteria, formerly known as Candidate Division TM7, represents a unique and enigmatic bacterial phylum with several distinctive characteristics:
- Ultra-small cell size (200-300 nm in diameter), significantly smaller than conventional bacteria
- Obligate epiparasitic lifestyle, growing on the surface of host bacteria
- Member of the Candidate Phyla Radiation (CPR), a large group of bacteria with reduced genomes and unusual properties
- Highly reduced genome (approximately 700-1,000 kilobases), lacking genes for many essential biosynthetic pathways
- Unable to synthesize amino acids, vitamins, or cell wall precursors, requiring these from host bacteria
- First identified in a peat bog in Germany (TM stands for "Torf, mittlere Schicht," German for "a middle layer of peat")
- Ubiquitous distribution across diverse environments including soil, seawater, wastewater, and animal-associated habitats
- Gram-positive cell wall structure, despite phylogenetic distance from other Gram-positive bacteria
- Lacks peptidoglycan biosynthesis genes but possesses a distinctive cell envelope
- Non-cultivable in axenic (pure) culture due to obligate dependence on host bacteria
- First oral strain (Nanosynbacter lyticus type strain TM7x, also known as HMT_952) was co-isolated with its bacterial host
- Displays a dynamic interaction with host bacteria, including potential host killing under certain conditions
- Remarkable 16S rRNA diversity with multiple subdivisions and clades
- Phylogenetically divided into at least 4 clades (2 monophyletic and 2 polyphyletic)
- High proportion (58%) of phylotypes are sample-specific, with few widely distributed members
- Most widely distributed and abundant phylotypes belong to subdivision 3
- Lacks genes for the tricarboxylic acid (TCA) cycle and electron transport chain
- Possesses genes for glycolysis and fermentation, suggesting a fermentative metabolism
- Contains genes for attachment to and interaction with host bacteria
- Exhibits a parasitic lifestyle that can range from commensalism to virulence depending on environmental conditions
- Possesses unusual ribosomal features, including self-splicing introns in some 16S rRNA genes
- Demonstrates high strain-level variation even within the same human host
- Exhibits host specificity, with different strains associating with different bacterial hosts
- Possesses genes for horizontal gene transfer, suggesting active genetic exchange with other bacteria
- Contains minimal translation, transcription, and replication machinery
- Lacks complete pathways for de novo nucleotide synthesis
- Possesses specialized surface proteins likely involved in host attachment and interaction
- Exhibits evolutionary adaptations consistent with genome reduction and host dependency
The unique biology of Candidatus Saccharibacteria represents a fascinating example of extreme genome reduction and symbiotic adaptation, providing insights into the minimal genetic requirements for bacterial life and the evolution of host-dependent lifestyles.
Role in Human Microbiome
Candidatus Saccharibacteria (TM7) occupies a unique niche within the human microbiome, primarily as an epiparasite of other bacteria rather than directly interacting with human cells. Its distribution and role across different body sites include:
Oral Cavity:
- Most prevalent and well-studied habitat for TM7 in humans
- Represents approximately 1% of the oral microbiome
- Present in multiple oral sites including dental plaque, saliva, tongue, and gingival tissue
- At least 6 distinct groups of TM7 have been identified in the oral cavity, varying in relative abundance across anatomic sites
- Particularly abundant in subgingival plaque and periodontal pockets
- Often associated with periodontal disease, with increased abundance in periodontitis patients
- The oral strain TM7x (Nanosynbacter lyticus) has been co-cultivated with its host bacterium Actinomyces odontolyticus
- Forms a dynamic relationship with host bacteria that can range from commensalism to parasitism
Gastrointestinal Tract:
- Present at lower abundance compared to the oral cavity
- Detected in fecal samples, suggesting colonization of the intestinal environment
- May interact with gut microbiota, potentially influencing community structure
- Specific roles in gut health and disease remain largely unexplored
- Potential association with inflammatory bowel disease has been suggested but requires further investigation
Urogenital Tract:
- Detected in vaginal samples at low prevalence
- Specific role in vaginal microbiome ecology is poorly understood
- May interact with other vaginal bacteria as epiparasites
- Potential implications for vaginal health remain to be determined
Skin:
- Present in the skin microbiome at low abundance
- Distribution across different skin sites and association with skin conditions remains poorly characterized
- Likely interacts with skin-associated bacteria as epiparasites
Other Body Sites:
- Surprisingly detected in some clinical samples including blood cultures and cardiac valves at very low prevalence (<3%)
- Presence in these normally sterile sites raises questions about potential translocation from primary colonization sites
- Role in clinical infections, if any, remains to be determined
Ecological Interactions:
- Functions primarily as an obligate epiparasite, growing on the surface of host bacteria
- Depends entirely on host bacteria for essential nutrients and metabolites
- Can modulate host bacterial physiology, potentially affecting growth dynamics
- May influence microbial community structure by affecting the abundance of host bacteria
- Under certain conditions, can exhibit virulent behavior, killing host bacteria
- Potentially affects oral microbial ecology by modulating the microbiome structure and functionality
Host Specificity:
- Different TM7 strains show specificity for different bacterial hosts
- In the oral cavity, primarily associated with Actinomyces, Streptococcus, and other common oral bacteria
- Host range and specificity in other body sites remain largely unexplored
- Strain-level variation observed even within the same human host
Prevalence Patterns:
- Highest prevalence in oral samples
- Moderate prevalence in fecal and breast milk samples
- Lower prevalence in vaginal and urine samples
- Very low prevalence in normally sterile sites (blood, cardiac valves)
- Prevalence patterns suggest primary ecological niches in the oral cavity and secondary colonization of other body sites
The role of Candidatus Saccharibacteria in the human microbiome represents a unique form of "microbe-on-microbe" interaction rather than direct microbe-host interaction. By parasitizing other bacteria, TM7 may indirectly influence human health through effects on microbial community structure and function. However, the full extent of its impact on human health and disease remains to be elucidated, highlighting the need for further research into this enigmatic bacterial phylum.
Health Implications
The health implications of Candidatus Saccharibacteria (TM7) are complex and not fully understood, primarily due to its unique lifestyle as an epiparasite of other bacteria rather than directly interacting with human cells. Current knowledge suggests several potential health associations:
Periodontal Disease:
- Increased abundance of TM7 has been observed in periodontal disease
- Multiple studies have reported higher levels of TM7 in subgingival plaque of periodontitis patients compared to healthy controls
- May contribute to disease by altering the abundance and behavior of other oral bacteria
- Could potentially modulate the virulence of host bacteria involved in periodontal pathogenesis
- The exact mechanism of TM7 contribution to periodontal disease remains unclear
- May serve as a potential biomarker for periodontal disease progression
Inflammatory Conditions:
- Preliminary evidence suggests possible associations with inflammatory bowel disease
- May influence inflammation through effects on the microbial community rather than direct interaction with the immune system
- Could potentially alter the balance of pro-inflammatory and anti-inflammatory bacteria
- The parasitic relationship with host bacteria might trigger changes in bacterial metabolism that influence inflammatory responses
- Research in this area remains limited and largely speculative
Indirect Effects on Host Immunity:
- As an epiparasite, TM7 may indirectly affect host immunity by modulating the behavior of host bacteria
- Could potentially alter the production of immunomodulatory compounds by host bacteria
- May influence biofilm formation and structure, affecting bacterial persistence and immune evasion
- The ultra-small size may allow TM7 to access niches not available to larger bacteria, potentially affecting microbial-immune interactions
Potential Role in Dysbiosis:
- May contribute to microbial dysbiosis by altering the abundance and function of key bacterial species
- The parasitic lifestyle could disrupt established microbial networks and community structures
- Under certain conditions, virulent behavior toward host bacteria could lead to ecological shifts in the microbiome
- These ecological changes might contribute to disease states characterized by microbial imbalance
Presence in Clinical Samples:
- Detected at low prevalence in normally sterile sites such as blood cultures and cardiac valves
- Unclear whether this represents contamination, translocation from primary sites, or potential involvement in infection
- No direct evidence currently links TM7 to invasive infections or systemic disease
- Further research is needed to determine the clinical significance of TM7 detection in these samples
Potential Therapeutic Implications:
- Understanding TM7-host bacterial interactions could provide novel approaches to microbiome modulation
- Targeting TM7 or its relationship with host bacteria might offer strategies for managing conditions associated with dysbiosis
- The obligate parasitic lifestyle presents unique vulnerabilities that could be exploited therapeutically
- Research into TM7 biology may reveal fundamental principles of bacterial interdependence relevant to microbiome-based therapies
Limitations in Current Understanding:
- Inability to culture most TM7 strains independently limits functional studies
- Reliance on molecular and imaging techniques provides incomplete information about physiological roles
- Causality versus correlation in disease associations remains difficult to establish
- Strain-level variation complicates interpretation of health associations
- The indirect nature of TM7's effects on human health makes mechanistic studies challenging
Future Research Directions:
- Development of improved co-culture systems to study TM7-host bacterial interactions
- Longitudinal studies to determine whether TM7 abundance changes precede or follow disease development
- Investigation of strain-specific effects on health and disease
- Exploration of potential metabolic interactions between TM7, host bacteria, and the human host
- Assessment of TM7's role in microbiome development and stability across the lifespan
The health implications of Candidatus Saccharibacteria highlight the complex ecological networks within the human microbiome. Rather than acting as a traditional pathogen or commensal, TM7 represents a "parasite of commensals" that may indirectly influence human health through effects on the microbial community. This unique ecological position underscores the importance of considering multi-level interactions within the microbiome when investigating microbial contributions to health and disease.
Metabolic Activities
The metabolic activities of Candidatus Saccharibacteria (TM7) are highly specialized and constrained by its ultra-small size and obligate epiparasitic lifestyle. Genomic analyses and limited cultivation studies have provided insights into its unique metabolic capabilities:
Reduced Metabolic Capacity:
- Extremely reduced genome (approximately 700-1,000 kilobases) with limited metabolic genes
- Lacks complete biosynthetic pathways for amino acids, nucleotides, fatty acids, and vitamins
- Absence of genes for the tricarboxylic acid (TCA) cycle
- Missing electron transport chain components for oxidative phosphorylation
- Lacks genes for de novo synthesis of cell wall precursors
- Minimal set of metabolic genes reflecting extreme genome reduction and host dependency
Energy Metabolism:
- Possesses complete or near-complete glycolytic pathway for glucose metabolism
- Relies primarily on fermentation for energy generation
- Contains genes for substrate-level phosphorylation to produce ATP
- Lacks respiratory chain components, suggesting an anaerobic or microaerophilic lifestyle
- May utilize alternative electron acceptors in some environments
- Energy metabolism appears streamlined for efficiency with minimal gene content
Nutrient Acquisition:
- Highly dependent on host bacteria for essential nutrients
- Possesses transporters for uptake of amino acids, nucleosides, and other essential compounds
- Contains genes for peptide and oligopeptide transport systems
- May obtain lipids and membrane components directly from host bacteria
- Likely scavenges metabolic intermediates from host bacterial metabolism
- Possesses specialized mechanisms for attachment to and nutrient extraction from host bacteria
Carbohydrate Metabolism:
- Contains genes for processing simple sugars
- Name "Saccharibacteria" reflects apparent specialization in sugar compound utilization
- May preferentially metabolize carbohydrates as carbon and energy sources
- Limited capacity for complex carbohydrate breakdown
- Likely depends on host bacteria or the human host for initial breakdown of complex carbohydrates
Protein and Amino Acid Metabolism:
- Lacks complete pathways for amino acid biosynthesis
- Contains genes for amino acid and peptide transporters
- Possesses some aminotransferases for amino acid interconversion
- May have li (Content truncated due to size limit. Use line ranges to read in chunks)