Klebsiella pneumoniae
Klebsiella pneumoniae is a gram-negative, encapsulated, non-motile bacterium belonging to the family Enterobacteriaceae and the phylum Proteobacteria. First isolated in the late 19th century and initially known as Friedlander's bacterium, K. pneumoniae has emerged as a significant pathogen of increasing clinical concern due to its ability to cause a wide range of infections and its growing resistance to antibiotics. In 2024, K. pneumoniae was designated as a Critical priority pathogen on the WHO Bacterial Priority Pathogens List—the highest category—emphasizing the urgent need for new antibiotics, vaccines, and infection control strategies.[1]
Key Characteristics
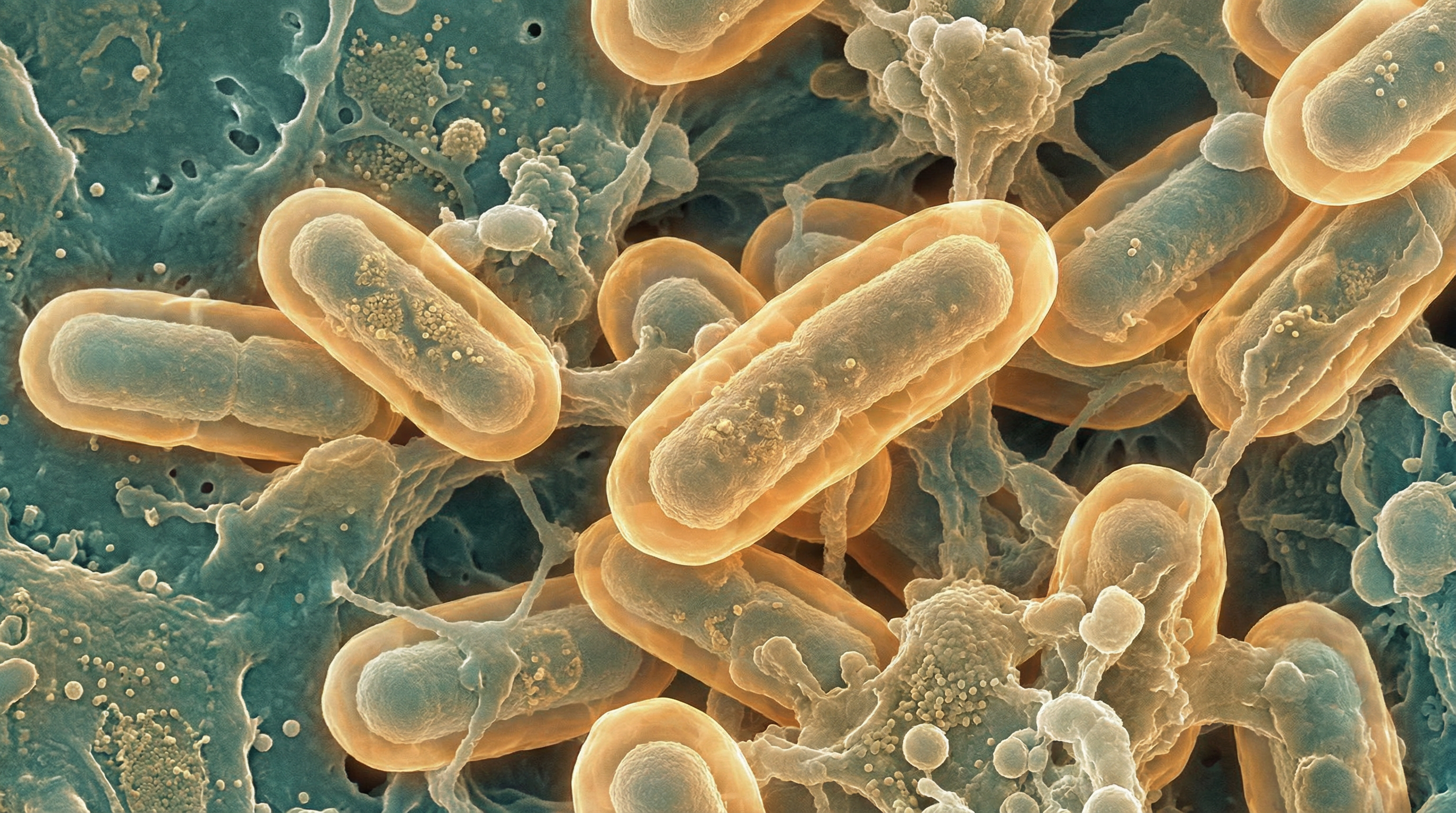
K. pneumoniae is a rod-shaped bacterium measuring approximately 0.3-1.0 μm in width and 0.6-6.0 μm in length. It is non-motile, facultatively anaerobic, and possesses a prominent polysaccharide capsule that gives colonies a mucoid appearance when grown on laboratory media. The bacterium typically forms large, dome-shaped colonies that are gray, moist, and sticky.
As a member of the Enterobacteriaceae family, K. pneumoniae is characterized by its ability to ferment glucose and other carbohydrates, reduce nitrate to nitrite, and test positive for catalase but negative for oxidase. It can utilize citrate as a sole carbon source and is generally urease-positive, which distinguishes it from some other members of the family.
K. pneumoniae is ubiquitous in the environment, found in soil, surface waters, plants, and on medical devices. It is also a common colonizer of the human gastrointestinal tract, where it typically exists as a commensal organism in healthy individuals. However, it can become an opportunistic pathogen under certain conditions, particularly in immunocompromised hosts or when the normal microbiota is disrupted, such as following antibiotic treatment.
The genome of K. pneumoniae is approximately 5-6 million base pairs in size and consists of a circular chromosome and often multiple plasmids. These plasmids frequently carry genes for antibiotic resistance and virulence factors, contributing to the pathogen's adaptability and clinical significance. Genomic studies have revealed considerable genetic diversity among K. pneumoniae strains, with different lineages associated with varying levels of virulence and antibiotic resistance.
Role in Human Microbiome
K. pneumoniae is primarily found in the human gastrointestinal tract, where it typically exists as a commensal organism in low abundance, constituting less than 1% of the gut microbiota in healthy individuals. Colonization rates vary significantly by geography: 5-35% in Western populations and 18.8-87.7% in Asian populations.[2] In a Chinese cohort study, 44.8% (69/154) of healthy individuals carried K. pneumoniae in their gut, with 5.1% harboring hypervirulent strains.[3]
Colonization Resistance and Dysbiosis
In the gut microbiome, K. pneumoniae occupies a niche alongside other Enterobacteriaceae, including Escherichia coli. Under normal conditions, its growth is kept in check by colonization resistance provided by the dominant anaerobic bacteria of the gut. This resistance arises from two key mechanisms:
- Exploitation competition: Commensal bacteria compete for nutrients, limiting resources available to K. pneumoniae
- Interference competition: Beneficial bacteria produce antimicrobial toxins that inhibit K. pneumoniae growth[4]
However, disruption of the gut microbiota, particularly through antibiotic use, significantly increases the risk of K. pneumoniae overgrowth. In a Cambodian NICU study, all common antibiotics except imipenem significantly increased acquisition risk: ampicillin-gentamicin (OR 1.96), ampicillin (OR 1.77), and ceftriaxone (OR 1.85).[5]
Type VI Secretion System for Gut Colonization
Recent research has revealed that K. pneumoniae employs a Type VI Secretion System (T6SS) to overcome microbiota-mediated colonization resistance. This molecular weapon system is critical for establishing gut colonization:
- 470 of 5,159 genes (9.11%) significantly contribute to colonization ability
- T6SS specifically targets and reduces Betaproteobacteria (Parasutterella, Burkholderiales)
- Expression of T6SS genes is 7-fold higher in the cecum compared to minimal medium
- T6SS is regulated by gut environmental cues including arginine, iron availability, and anaerobic conditions[4]
Gut as Reservoir for Infection
The gut serves as a major reservoir for subsequent K. pneumoniae infections, particularly in healthcare settings. Prior gut colonization is the primary risk factor for K. pneumoniae infection in ICU patients. Importantly, gut hypervirulent K. pneumoniae strains are phylogenetically closely related to fatal clinical strains—71.64% of gut-clinical strain pairs had <1000 SNPs (range 118-520), suggesting gut colonization directly leads to invasive community-acquired infections.[3]
Notably, healthy human gut is NOT a major reservoir of carbapenem-resistant K. pneumoniae or a hotspot for convergence of carbapenem-resistance and virulence genes—none of the gut hvKp isolates in Chinese studies carried carbapenem resistance genes.
Health Implications
K. pneumoniae is responsible for 12.3% of healthcare-associated infections in Europe and causes a wide range of infections, from relatively mild to life-threatening.[1] Its health implications can be categorized based on the infection site and severity:
Respiratory Infections
K. pneumoniae causes classic Friedlander's pneumonia with high mortality rates:
- Pre-antibiotic era mortality: ~80%
- Bacteremic community-acquired pneumonia (hvKp): 55.1% mortality
- Community-acquired empyema (hvKp): 32.4% mortality[2]
Clinical features include rapid onset with high fever, production of thick bloody sputum ("currant jelly"), and tendency to cause necrosis, cavitation, and abscess formation. Hypervirulent strains can cause severe pneumonia even in previously healthy individuals.
Urinary Tract Infections (UTIs)
UTIs represent 48.5% of clinical K. pneumoniae isolates, making this the most common source of infection.[6] These infections range from uncomplicated cystitis to complicated pyelonephritis and are associated with:
- Urinary catheterization
- Structural abnormalities of the urinary tract
- Recurrent infections due to biofilm formation (82% of isolates form biofilms)
- Treatment failure due to antibiotic resistance
Bloodstream Infections (Bacteremia)
K. pneumoniae bacteremia accounts for 11.0% of clinical sources and carries high mortality, particularly in carbapenem-resistant strains (30-70% mortality).[1] Independent predictors of mortality include:
- Charlson Comorbidity Index
- Chronic kidney disease
- Recent renal replacement therapy
- Acute kidney injury following bloodstream infection
- Septic shock and delayed appropriate therapy
Pyogenic Liver Abscesses
A distinctive syndrome of community-acquired pyogenic liver abscess (PLA) is the hallmark of hypervirulent K. pneumoniae. This syndrome, initially described in East Asia but now global, is characterized by:
- Community-acquired infection in otherwise healthy individuals
- Metastatic spread to spleen, eyes (endophthalmitis), and meninges
- Association with K1 and K2 capsular serotypes (>70% of hvKp infections)
- High virulence: hypervirulent strains are ~100,000-fold more virulent than classical strains in animal models (LD50 ~10³ CFU vs ~10⁸ CFU)[6]
Endophthalmitis
Metastatic endophthalmitis occurs in approximately 5% of hvKp bacteremia cases and has devastating outcomes:
- 89% of cases result in light perception or worse
- 41% of affected eyes require evisceration or enucleation[2]
Necrotizing Fasciitis
K. pneumoniae necrotizing fasciitis has 47% mortality—significantly higher than group A streptococcus (19%).[2]
Other Infections
K. pneumoniae can cause various other infections, including:
- Meningitis: Particularly in neonates and post-neurosurgical patients; hvKp meningitis has worse outcomes than S. pneumoniae
- Wound infections: 9.5% of clinical sources
- Osteomyelitis, endocarditis, and intra-abdominal infections
Antibiotic Resistance
One of the most concerning health implications of K. pneumoniae is its increasing resistance to antibiotics. The WHO has designated carbapenem-resistant K. pneumoniae (CRKP) as the #1 Critical priority pathogen requiring urgent attention.
Extended-Spectrum Beta-Lactamases (ESBLs)
Over 1.5 billion people globally are colonized with ESBL-producing Enterobacteriaceae. ESBL prevalence varies by region:
- US collections: 78.6% (2,132/2,708) contain ESBL genes
- India: 70-90% of Enterobacteriaceae are ESBL-positive
- Cambodia NICU: 99% (1,412/1,423) of 3GC-R K. pneumoniae were ESBL producers
The dominant genotypes include CTX-M-15 (most common globally), CTX-M-14, SHV, and TEM.[7]
Carbapenemases
Three major classes of carbapenemases limit treatment options:
| Enzyme | US Prevalence | Coverage |
|---|---|---|
| KPC | 86% of CRE | Hydrolyzes all beta-lactams |
| NDM | 9% of CRE | Hydrolyzes all except aztreonam |
| OXA-48 | 4% of CRE | Primarily carbapenems, weak cephalosporin activity |
KPC resistance to ceftazidime-avibactam occurs in 10.5% of KPC-Kp bloodstream infections, with 22% in-hospital mortality. Notably, >60% of these strains regain susceptibility to at least one carbapenem due to mutations that restore carbapenem activity while conferring ceftazidime-avibactam resistance.[8]
Mortality Rates
- CRKP overall: 30-70% mortality
- Hypervirulent CRKP (hv-CRKP): 42.3% mortality (1.48-fold higher than classical CRKP)
- 18.2% of all CRKP isolates worldwide are hypervirulent[9]
High-Risk Clones
Specific sequence types facilitate global dissemination of resistance:
- ST258: 40.6% of convergent (MDR + hvKp) isolates in US
- ST307: 22.1% of convergent isolates in US
- ST11: Dominant in Asia, associated with fatal outbreaks
- ST147 and ST101: Additional high-risk lineages[6]
Future Projections
Deaths attributable to carbapenem-resistant Gram-negative bacteria increased from 127,000 in 1990 to 226,000 in 2021 (78% increase). By 2050, antimicrobial resistance is projected to cause 1.91 million attributable deaths annually with 39.1 million cumulative deaths between 2025-2050.[10]
Hypervirulent Strains
In recent decades, hypervirulent strains of K. pneumoniae (hvKp) have emerged, primarily in East Asia but now spreading globally. These strains are defined by key characteristics:
Defining Features
- Hypermucoviscosity phenotype: Positive string test (>5 mm) in ~90% of clinical hvKp strains
- 6-10 fold increase in siderophore production compared to classical strains
- Dominant capsular serotypes: K1 and K2 account for >70% of hvKp infections; also K5, K20, K54, K57
- ~100,000-fold more virulent in animal models (LD50 ~10³ CFU vs ~10⁸ CFU for classical strains)[11]
Key Virulence Factors
Aerobactin is the dominant siderophore and most critical virulence factor:
- Accounts for ~90% of total siderophore production in hvKp
- Present in >90% of hvKp strains vs only ~6% of classical strains
- Production levels: hvKp produces 138 μg/ml vs 23.1 μg/ml in classical strains
- iucA deletion decreases siderophore production by 92-96% and significantly increases animal survival[12]
Capsule provides protection against phagocytosis and complement-mediated killing. As many as 186 KL types have been identified genomically, though traditional serotyping recognizes at least 79 distinct serotypes.
Convergent Strains: The Ultimate Threat
The emergence of strains combining multidrug resistance with hypervirulence represents a critical public health threat:
- 1.8% of US isolates (47/2,608) are convergent strains
- ST258 (40.6%) and ST307 (22.1%) are dominant convergent lineages
- Important finding: Most convergent isolates show unexpectedly low virulence despite possessing virulence genes, suggesting gene presence alone doesn't guarantee hypervirulence[6]
Nosocomial Shift
While hvKp was initially community-acquired, >90% of hvKp-causing infections are now nosocomial in Beijing retrospective studies, indicating a significant epidemiological shift.[3]
Metabolic Activities
K. pneumoniae exhibits versatile metabolic capabilities that contribute to its success as both a commensal and a pathogen:
Carbohydrate Metabolism
K. pneumoniae can utilize a wide range of carbohydrates as carbon sources, including:
Glucose: Metabolized through glycolysis and the tricarboxylic acid (TCA) cycle under aerobic conditions, or through mixed acid fermentation under anaerobic conditions.
Lactose: Unlike some other Enterobacteriaceae, K. pneumoniae can ferment lactose, producing acid and gas.
Complex carbohydrates: Some strains can metabolize complex plant-derived carbohydrates, reflecting the bacterium's environmental niche in soil and on plants.
L-fucose: Recent research has shown that K. pneumoniae can metabolize L-fucose, a sugar abundant in the gut mucus layer, providing a competitive advantage during gut colonization.
Nitrogen Metabolism
K. pneumoniae has several pathways for nitrogen acquisition and metabolism:
Nitrogen fixation: Some strains possess the ability to fix atmospheric nitrogen, a relatively rare trait among human-associated bacteria.
Urease activity: K. pneumoniae produces urease, which hydrolyzes urea to ammonia and carbon dioxide. This not only provides a nitrogen source but also creates a more alkaline microenvironment that can protect the bacterium from acid stress.
Amino acid catabolism: The bacterium can utilize various amino acids as both carbon and nitrogen sources.
Respiration and Fermentation
As a facultative anaerobe, K. pneumoniae can adapt its energy metabolism to different oxygen conditions:
Aerobic respiration: Under oxygen-rich conditions, K. pneumoniae uses oxygen as the terminal electron acceptor in its respiratory chain.
Anaerobic respiration: In the absence of oxygen, it can use alternative electron acceptors such as nitrate, fumarate, or dimethyl sulfoxide.
Fermentation: Under strictly anaerobic conditions without alternative electron acceptors, K. pneumoniae can ferment carbohydrates, producing a mixture of acids, alcohols, and gases.
Iron Acquisition
Iron is an essential nutrient for K. pneumoniae, and the bacterium has evolved sophisticated mechanisms for iron acquisition, particularly important during infection when iron is limited:
Siderophores: K. pneumoniae produces multiple siderophores, including enterobactin, yersiniabactin, aerobactin, and salmochelin. These high-affinity iron-binding molecules sequester iron from the environment or host proteins and transport it into the bacterial cell.
Heme utilization: Some strains can utilize heme as an iron source, extracting iron from host hemoproteins.
Ferrous iron transport: Under anaerobic or microaerobic conditions, K. pneumoniae can directly transport ferrous iron (Fe²⁺) through specific membrane transporters.
Hypervirulent strains often produce multiple siderophores simultaneously, enhancing their ability to acquire iron during infection and contributing to their increased virulence.
Biofilm Formation
K. pneumoniae can form biofilms, structured communities of bacteria embedded in a self-produced matrix of extracellular polymeric substances. Biofilm formation involves complex metabolic adaptations:
Exopolysaccharide production: The bacterium produces extracellular polysaccharides that form the biofilm matrix.
Metabolic heterogeneity: Bacteria within biofilms exhibit different metabolic states depending on their position, with those deeper in the biofilm often adopting a more dormant state.
Quorum sensing: K. pneumoniae uses quorum sensing systems to coordinate gene expression based on population density, influencing biofilm development.
Biofilms contribute to K. pneumoniae's persistence on medical devices and in chronic infections, and provide protection against antibiotics and host immune defenses.
Clinical Relevance
The clinical relevance of K. pneumoniae spans diagnosis, treatment, prevention, and infection control:
Diagnosis
Diagnosing K. pneumoniae infections typically involves:
Culture-based methods: Isolation of the bacterium from clinical specimens (blood, urine, sputum, wound exudates) on selective and differential media. K. pneumoniae forms large, mucoid colonies on media such as MacConkey agar, where it appears as lactose-fermenting (pink) colonies.
Biochemical identification: Traditional biochemical tests or automated systems to identify the species based on metabolic characteristics.
Molecular methods: PCR-based assays for rapid identification, particularly useful for detecting specific resistance genes or virulence factors.
Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS): Provides rapid species identification based on protein profiles.
Antimicrobial susceptibility testing: Essential for guiding appropriate antibiotic therapy, particularly given the high prevalence of resistance.
String test: A simple test for the hypermucoviscosity phenotype associated with hypervirulent strains, where a bacterial colony is stretched with a loop, forming a string >5 mm in length.
Molecular typing: Methods such as multilocus sequence typing (MLST), whole-genome sequencing, and capsular typing to identify specific lineages or clones, particularly important for outbreak investigations.
Treatment
Treatment of K. pneumoniae infections has become increasingly challenging due to antibiotic resistance:
- Antibiotic therapy: The choice of antibiotics dep (Content truncated due to size limit. Use line ranges to read in chunks)